Link to publication: Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling
Organisms from bacteria to animals and plants must protect themselves against pathogens. Homologous protein motifs are present in the immune pathways of all these organisms. One of these motifs is the TIR (Toll/interleukin-1 receptor/resistance protein) domain, which is named after the mammalian and plant immune receptors.
The best-known function of TIR proteins is signalosome assembly. However, recent work revealed that many TIR proteins have enzymatic functions, most commonly cleaving NAD+ (the oxidised form of nicotinamide adenine dinucleotide). NAD+ is a metabolite with redox properties that has roles in many cellular processes. Both signalosome assembly and enzyme functions depend on self-association into filamentous, open-ended complexes. TIR domains cleave NAD+ into nicotinamide and ADPR (adenosine diphosphate-ribose), but often also convert ADPR into its cyclic form (cADPR). Using NMR and crystallography, we found that bacterial TIR proteins can produce at least two new cADPR isomers. These are cyclised though an O-glycosidic bond; based on the location of the bond between the ribose moieties in ADPR, we named them 2′cADPR and 3′cADPR. 3’cADPR activates a protein from bacterial anti-phage defence system by allosterically modulating its oligomeric structure. The same molecule also acts as an inhibitor of the plant immune system. While functions of 2‘cADPR in bacteria are currently unknown, the same molecule is produced by plant immune receptors and may be an intermediate in the production of related nucleotides that activate the plant immune system (Fig. 1).
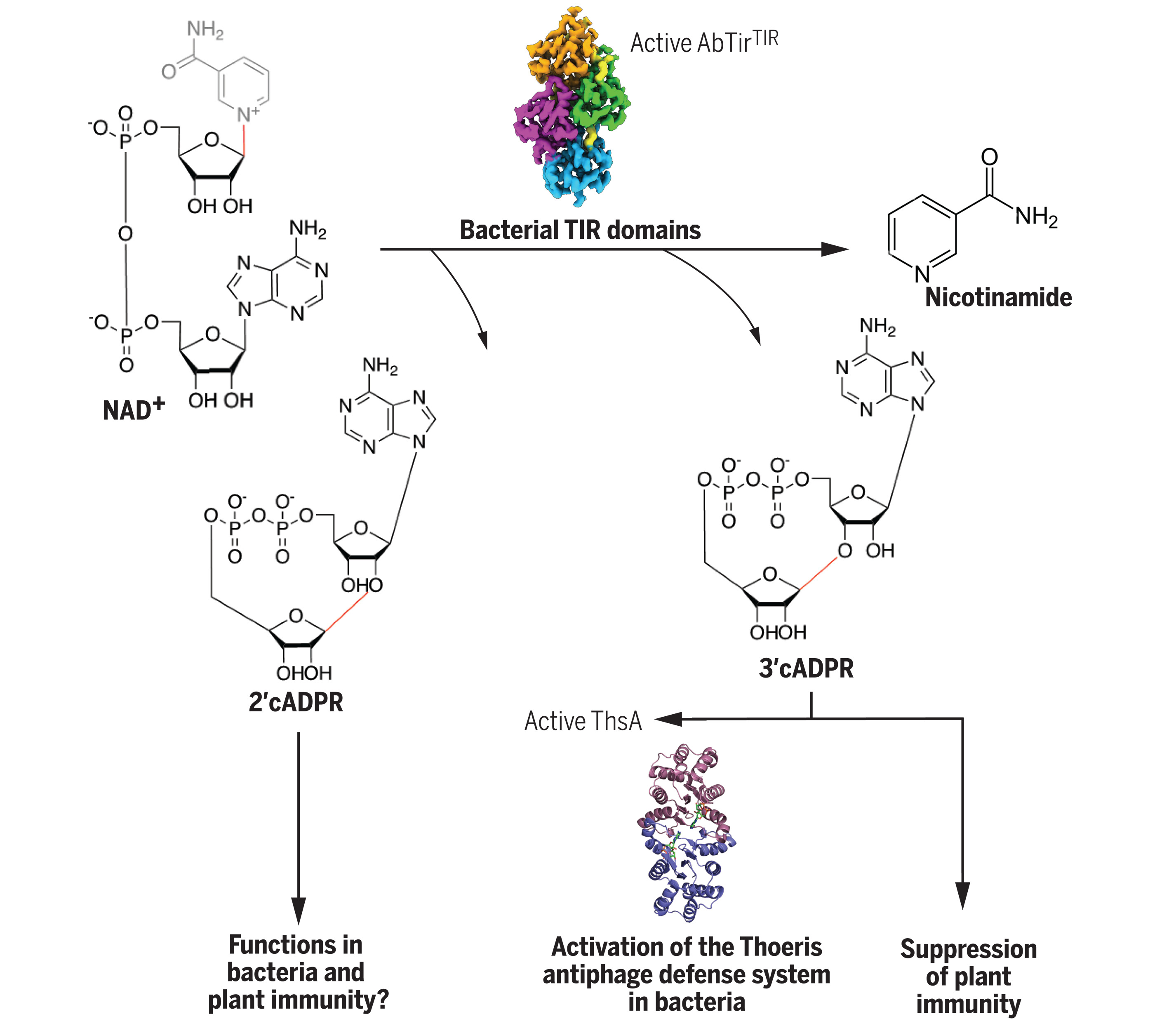
Figure 1. Overview of the study characterizing diverse immune roles of bacterial cADPR isomers. Bacteria have TIR domain-containing proteins that cleave NAD+ to produce cyclic ADPR isomers with different cyclic linkages. One of these molecules, 3’cADPR, has different roles in immunity pathways - it acts as an activator of the Thoeris anti-phage defence systems, by binding to the protein called ThsA. When produced by the effector HopAM1 from the plant pathogen Pseudomonas syringae, it suppresses plant immunity.
We used a stable NAD+ mimic 3AD (8-amino-isoquinoline adenine dinucleotide) to capture the active form of the assembly of TIR domains from a protein from Acinetobacter baumannii that produces 2’cADPR, and determined the structure by cryoEM: After incubation of the protein sample and 3AD, we observed some filaments under negative-stain TEM. Then we optimized the grids and freezing conditions for cyoEM data collection. We tried to decrease the protein concentration as the filaments were overlaying (Fig. 2b). However, the optimization of protein concentration was not successful, as we could not avoid the thick-ice problem. Therefore, we used Quantifoil Au R 1.2/1.3 300 mesh holey carbon girds and glow-discharged for 30 s at medium level on both sides. The we added 2 μL sample solution to the grids. Samples were vitrified in a Leica EMGP2 plunge freezer using a blotting time of 8.5 s at 8°C, with 96% humidity.
Although half of the grid was covered with ice (Fig. 2a), we used the other half to collect around 2000 movies using JEOL Cryo-ARM300 operated at 300 keV and equipped with an in-column Ω energy filter (slit width 20 eV) and Gatan K3 direct electron detector. All data processing was performed with cryoSPARC (Fig. 3). ChimeraX, ISOLDE, Coot and Phenix were used for model building and refinement, yielding the final structure of AbTirTIR:3AD filaments at 2.74 Å resolution.
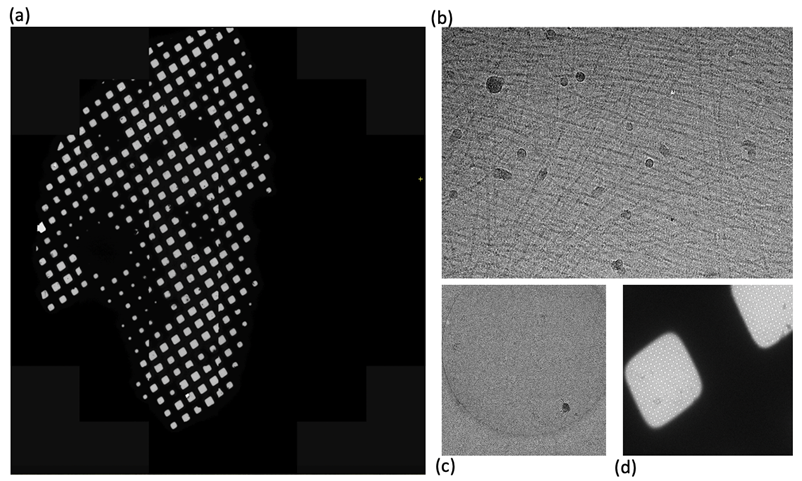
Figure 2. Data collection by cryo-EM. (a-d) Cryo-EM micrographs by ARM-300 under different magnifications.
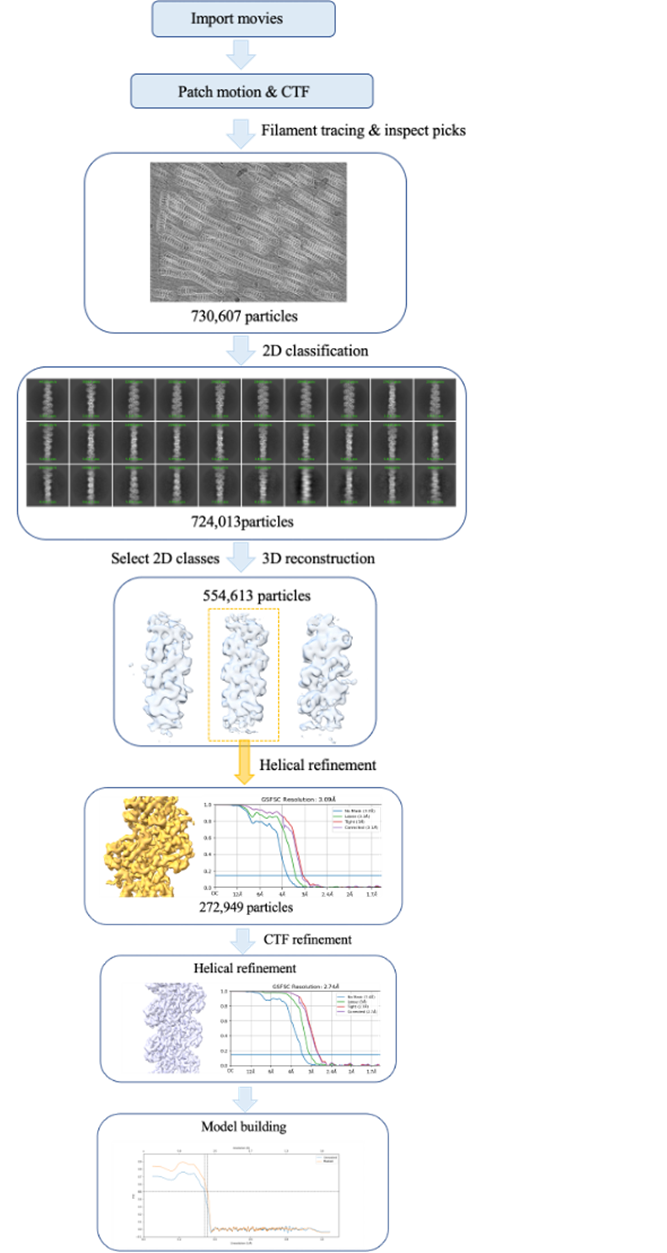
Figure 3. Workflow of data processing.
The cryoEM structure shows the presence of the 3AD molecule between two monomers of AbTirTIR. The filament is hold together by of two interfaces. The intrastrand “BE interface” involves the “BB-loop” (an important functional feature in TIR domains), while the interstrand “BCD interfaces” involve one molecule interacting with two molecules in the parallel strand (Fig. 4a). Incredibly, the assembly is a filament structurally analogous to those formed by animal TIR domains involved in Toll-like receptor signalling like MAL, which contains two parallel strands of TIR domains arranged head-to-tail (Fig. 4b), but different from enzyme TIR protein assemblies such as those from SARM1 and plant TIRs.
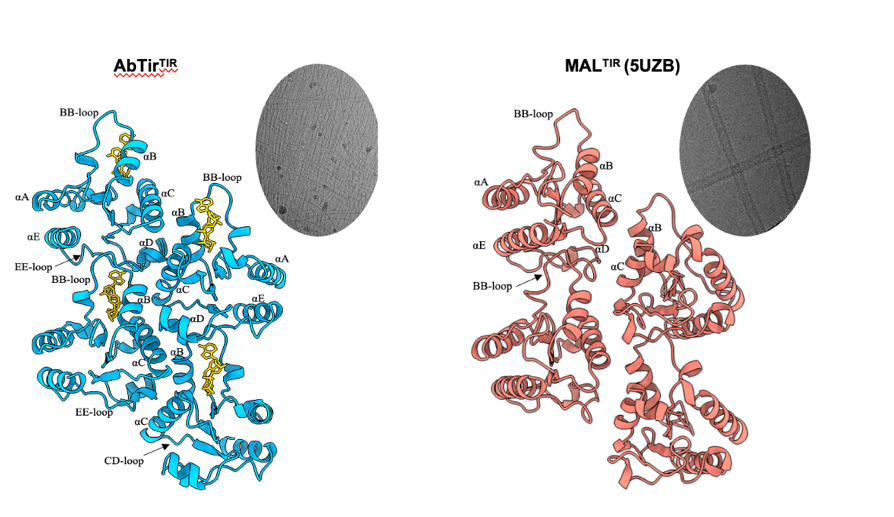
Figure 4. Similarity of filaments formed by the TIR domains from AbTir and the Toll-like receptor adaptor MAL.
Comparing cryoEM structure with the crystal structure of the monomeric domain, the filament assembly formation is accompanied by large conformational changes involving the BB-loop and the B and C helices (Fig. 5). The BB-loop and B helix have tilted outwards by ~50-55, while the C helix has refolded and includes residues from both the CC and CD loops.
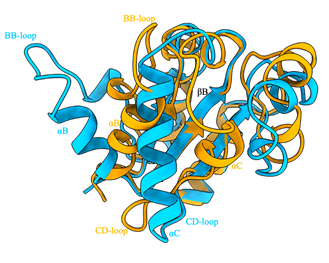
Figure 5. Structural comparison of AbTirTIR ctystal and cryoEM structures. Cyan: crystal structure; orange: cryoEM structure. The BB-loop and B helix have tilted outwards by ~50-55 while the C helix has refolded and includes residues from both the CC and CD loops.
In conclusion, our study reveals the chemical structures of new cADPR isoforms with signalling functions and provides information on how they are produced by TIR domains and how they signal in the immune pathways of bacteria and plants. Future studies will define how additional domains in bacterial TIR proteins facilitate the active configuration and what size active complexes form in bacterial cells.
